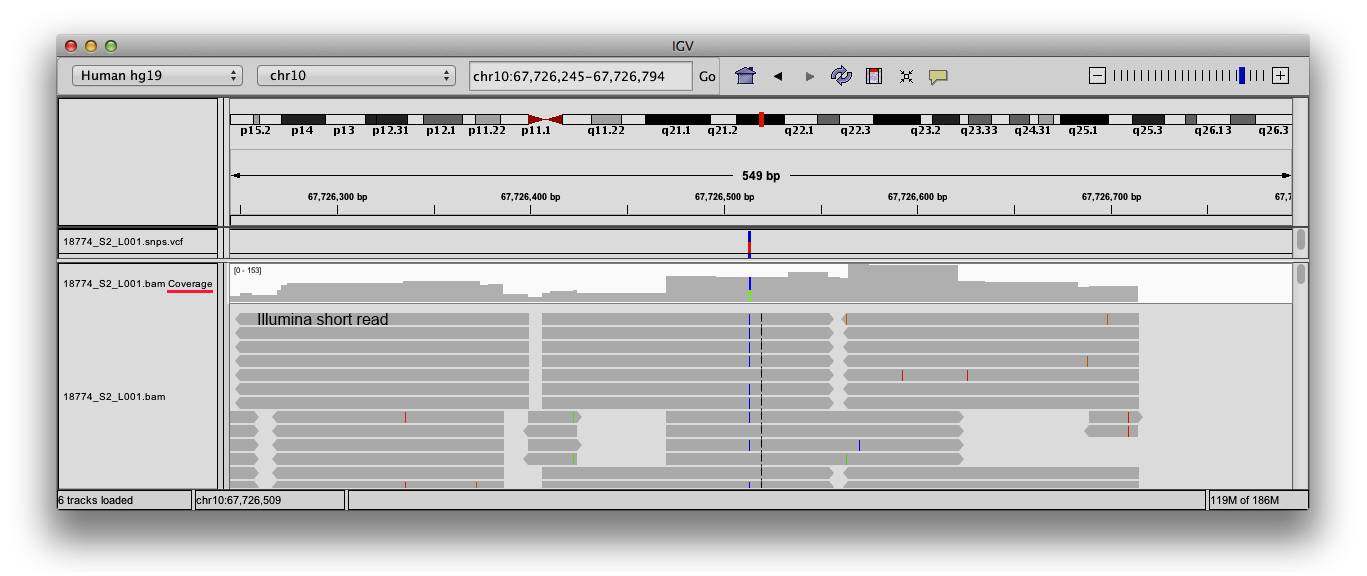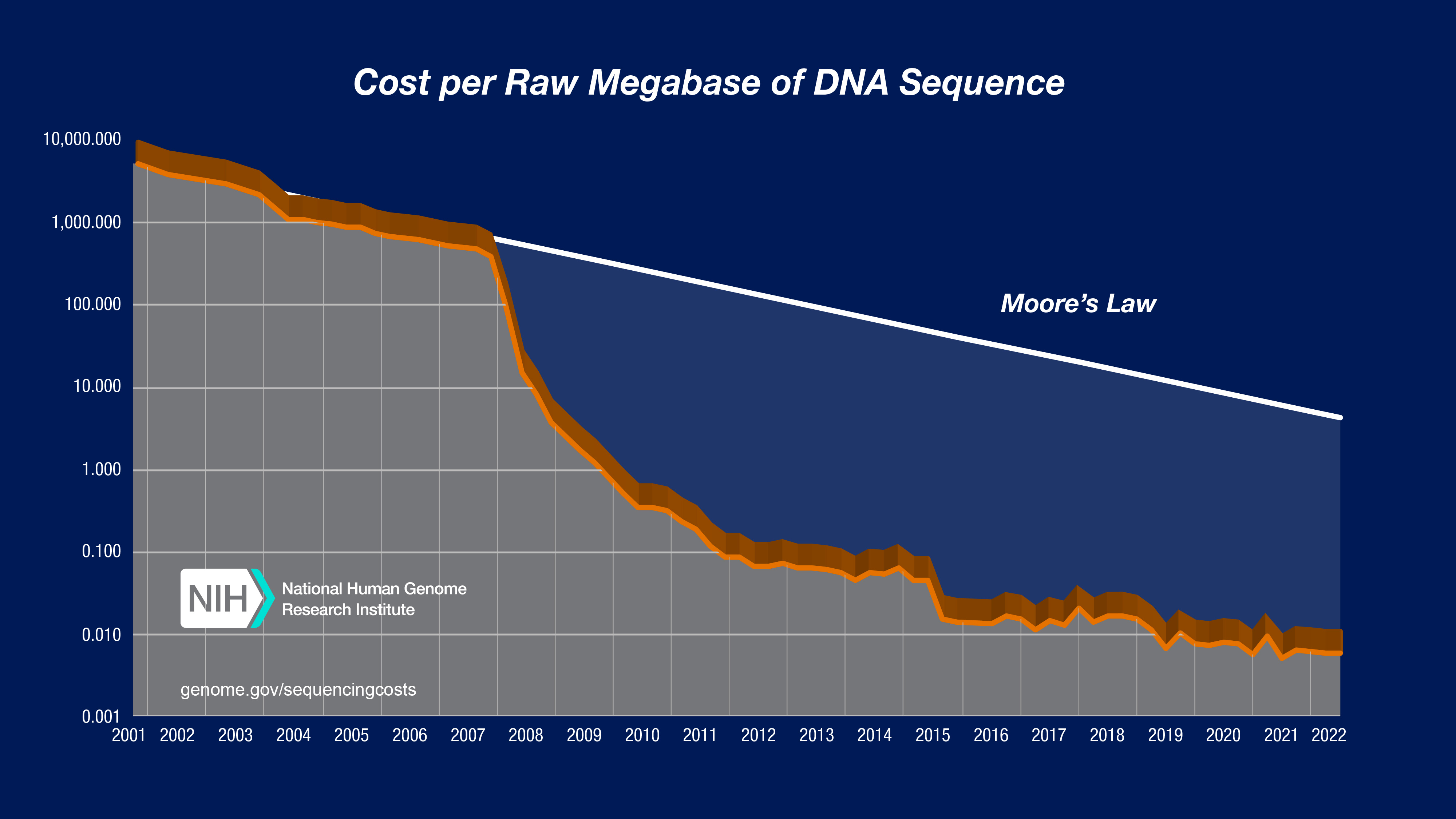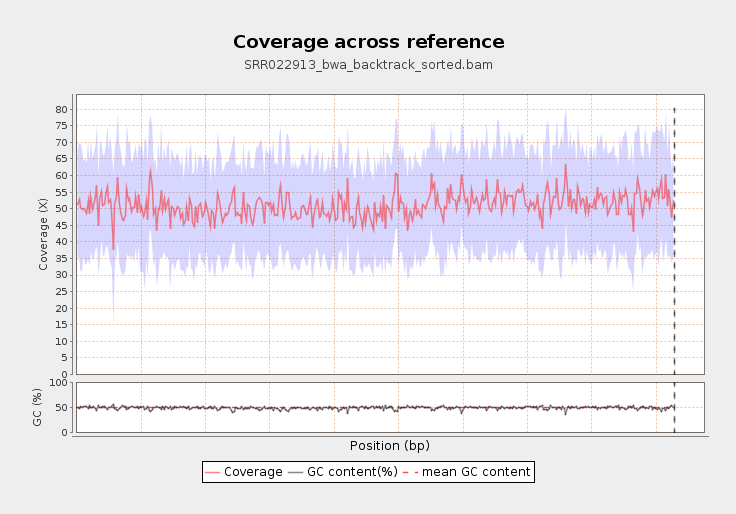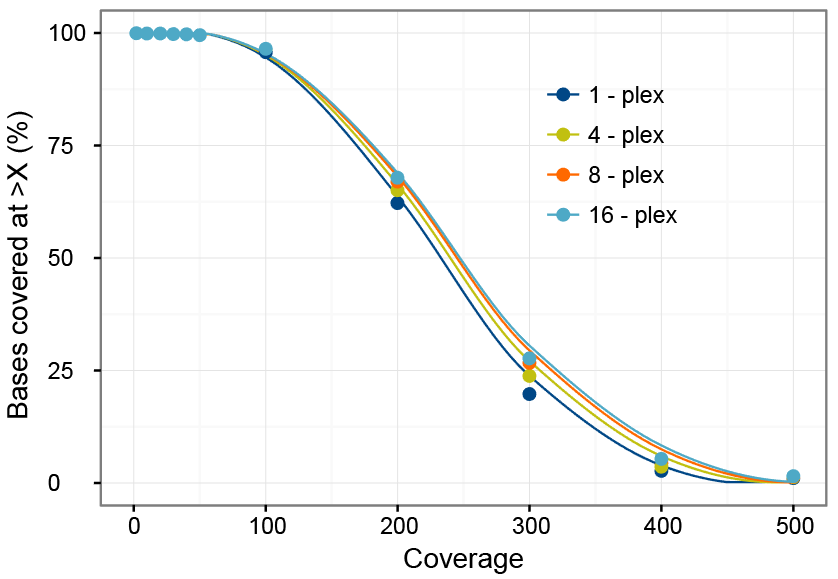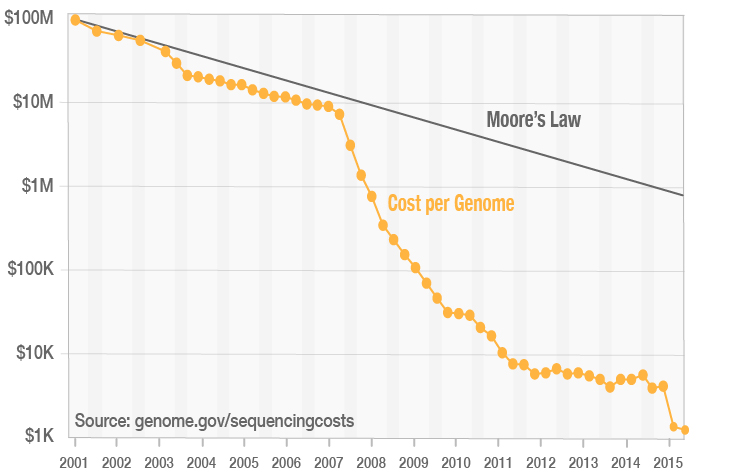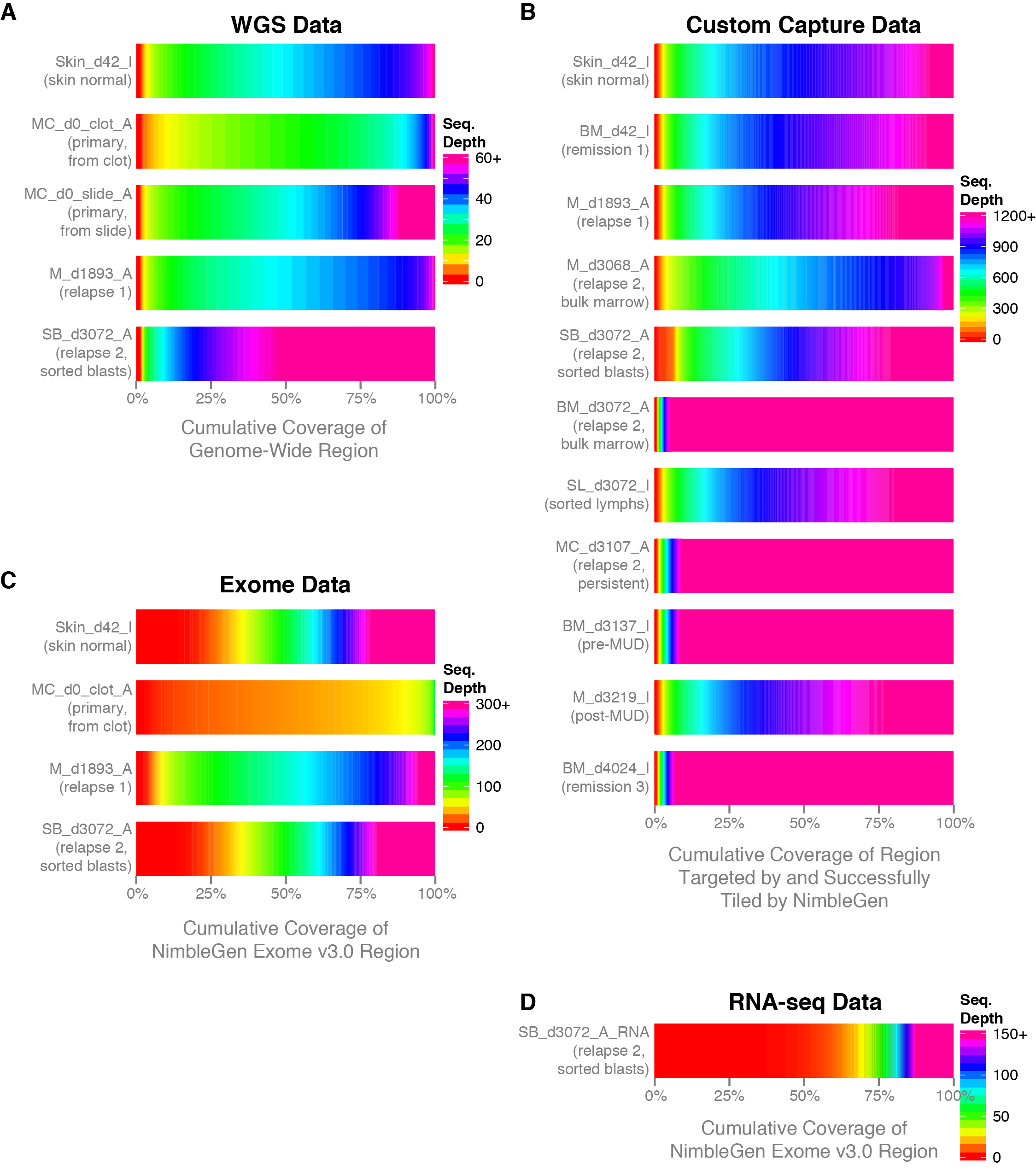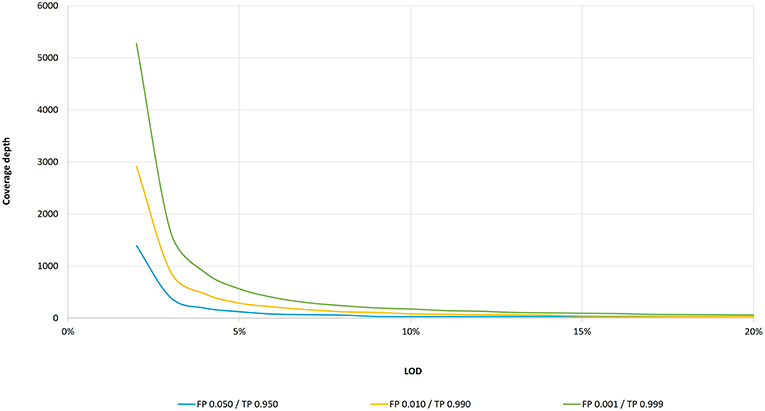
Frontiers | Standardization of Sequencing Coverage Depth in NGS: Recommendation for Detection of Clonal and Subclonal Mutations in Cancer Diagnostics

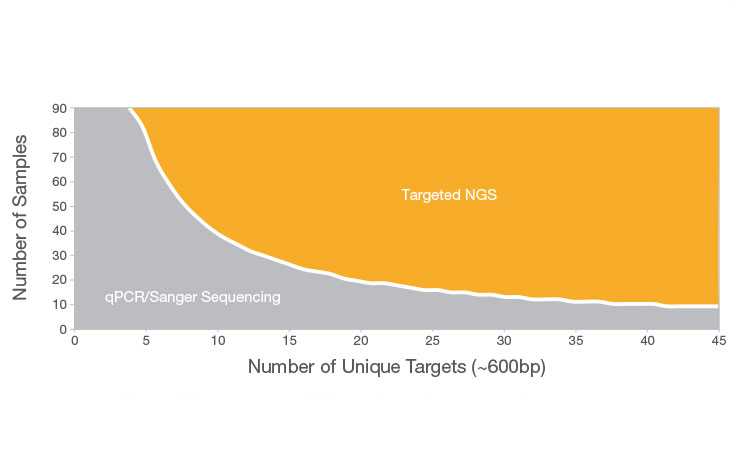
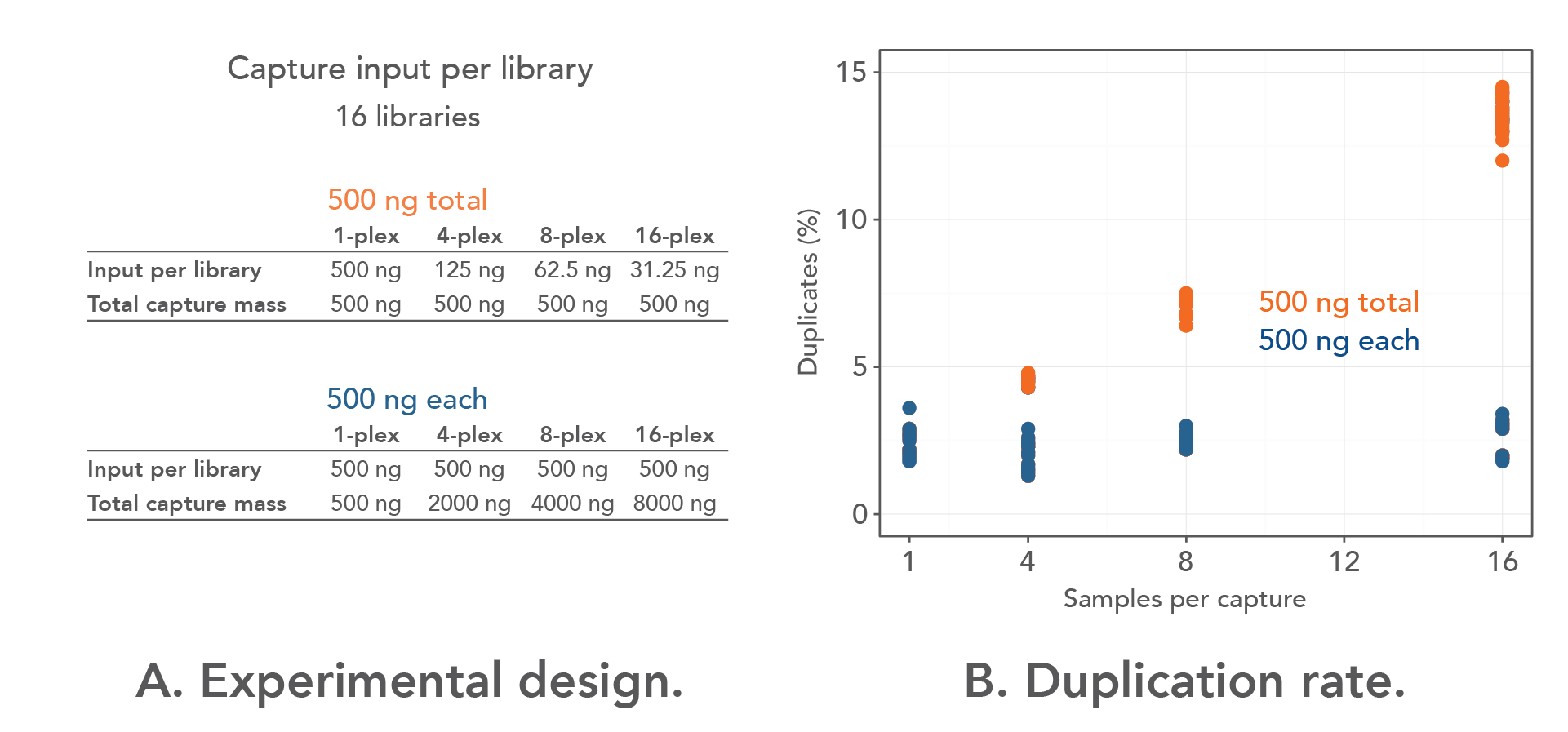
Calculation of contig number for various combination of c, L and T, by... | Download Scientific Diagram

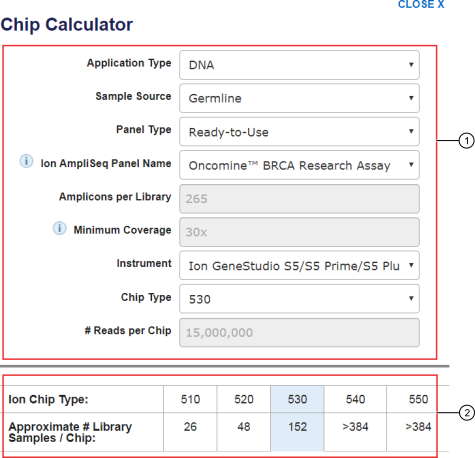
Devyser on X: "Did you know we have a Coverage Calculator which can help your sequencing planning? Just select your system and kit, the number and type of samples, and easily calculate

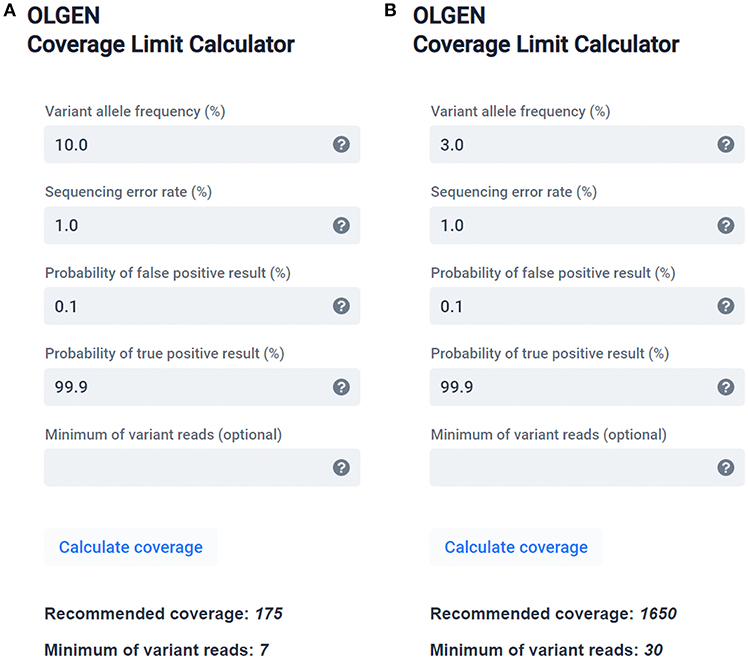
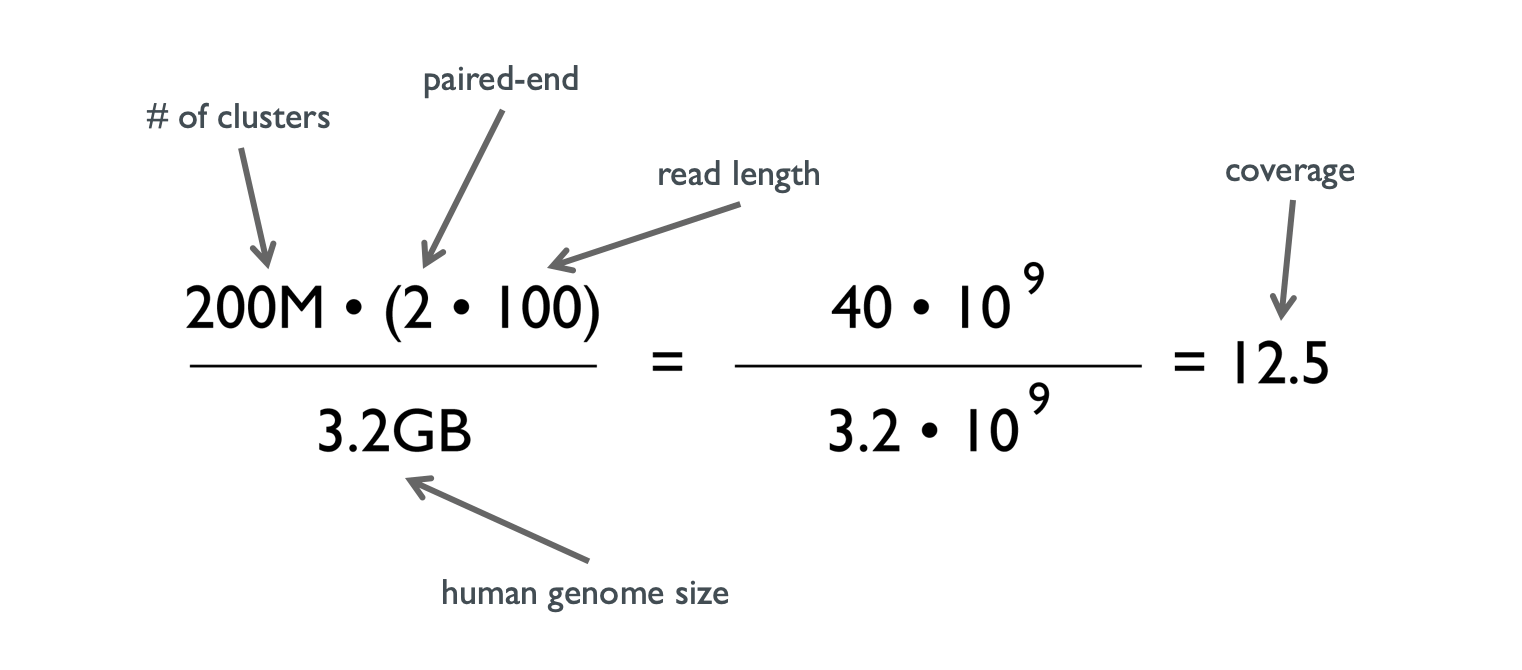
Mathematical Framework Provides a Read Depth Calculator and Guidelines... | Download Scientific Diagram

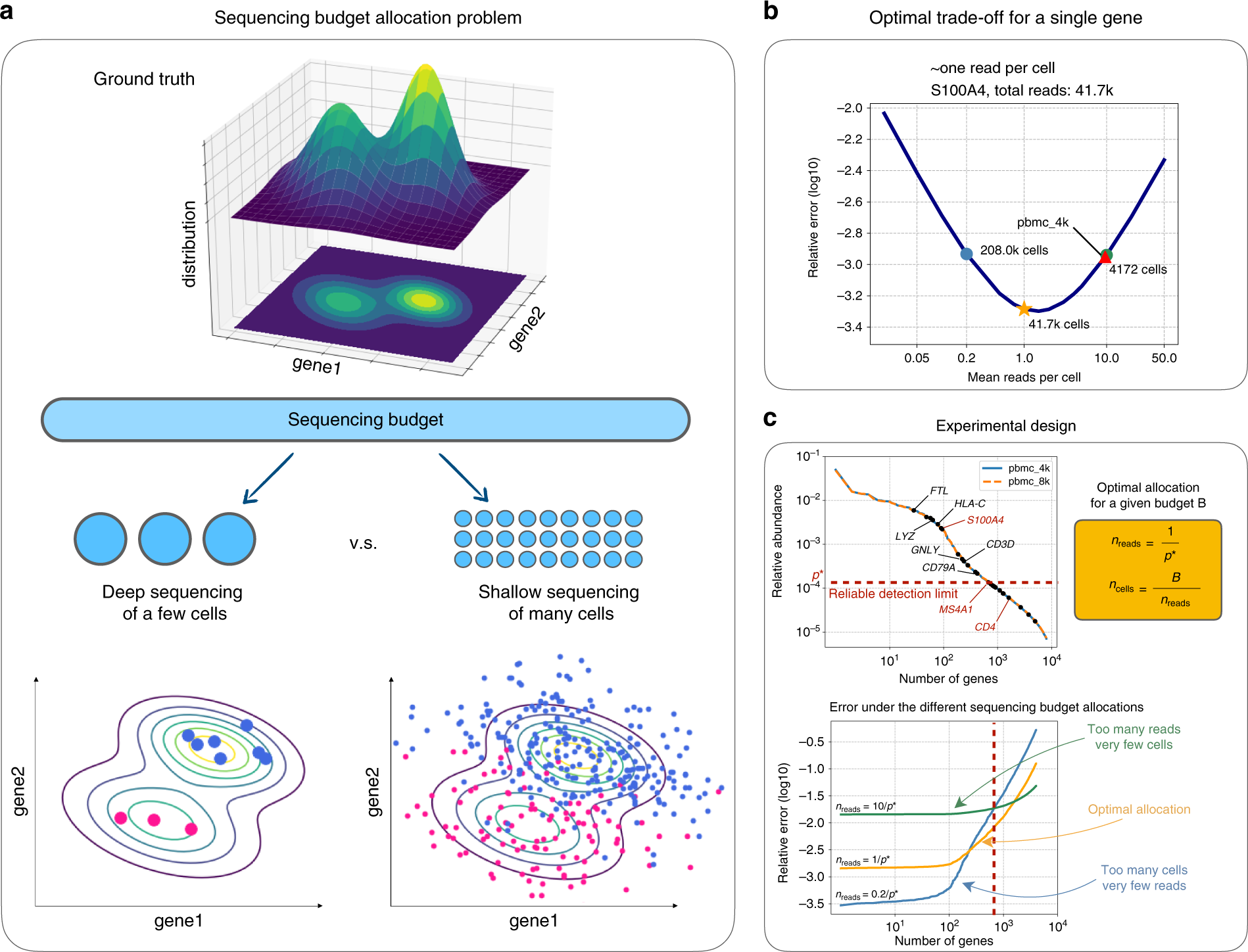
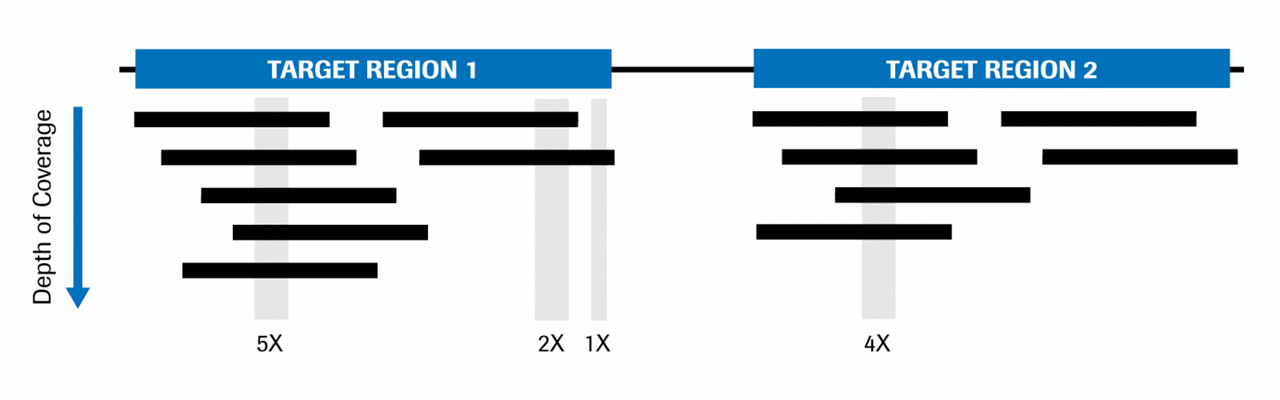
Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments | R-bloggers